News > Pathoplexus November Update
Pathoplexus November Update
By the Pathoplexus Team - 18 November 2025
Since our last update in August, Pathoplexus has continued to grow - both in data and in community. Over the past two months, thousands of new sequences have been shared, several new features have gone live, and new members have joined our global community! In addition, we were excited to be part of the joint PHA4GE & IPSN Conference - read on to find out more!
Data and Pathogen Updates
Pathoplexus now hosts over 8,900 directly-submitted sequences, including new uploads of:
-
1,366 new mpox sequences, from Pathogen Genomic Lab, DRC; INSA, Portugal; National Virus Reference Laboratory, Ireland; Laboratoire de Biologie moléculaire et d’Immunologie, Togo; Public Health Agency of Canada, Canada; Institute Pasteur de Dakar, Senegal; National Public Health Agency & Sierra Leone Ministry of Health, with Broad and Scripps Institute and with Institute Pasteur, Senegal and CDC, Sierra Leone; Institute National de Recherche Biomédicale, DRC; and National Genomic Sequencing Reference Laboratory, Malawi
-
7 new West-Nile sequences, from Infectious & Tropical Diseases Unit, Annunziata Hospital, Italy and Unité des Virus Émergents, Aix-Marseille Université, France
-
68 new Ebola-Zaire sequences, including those from past samples and from the recent outbreak, from Institut National de Recherche Biomédicale, DRC
-
1,326 new RSV-A sequences, from Grubaugh Lab, Yale, USA; Boston University & Boston Medical Center, Massachusetts, USA; WHO Influenza Collaborating Center, Australia (including samples from the Philippines); and Computational Evolution Group, ETH Zurich, Switzerland
-
1,642 new RSV-B sequences, from Grubaugh Lab, Yale, USA; Boston University & Boston Medical Center, Massachusetts, USA; and WHO Influenza Collaborating Center, Australia (including samples from the Philippines)
As well as directly uploaded sequences, we also ingest data from INSDC databases, ensuring users can access all openly available sequences via Pathoplexus.
A note on RSV
We have added 2,325 RSV-A and 1,335 RSV-B additional sequences from INSDC that are misclassified in NCBI taxonomy (under bovine orthopneumovirus) and were, hence, previously missed by our pipeline.
Click for more details on the RSV ingest update
To help ensure we could capture all available RSV sequences, we recently updated our RSV ‘minimizer’ (used to classify sequences). Previously we were ingesting from taxon ID 11250, which is annotated as “human respiratory syncytial virus”. We noticed that some human sequences have been annotated with taxon ID 12814 (“respiratory syncytial virus”), which is currently listed in NCBI Taxonomy as a child of “Bovine orthopneumovirus” (taxon ID 11246) and not under 11250. By updating the classifier we were able to ingest from the full set of sequences under Orthopneumovirus (taxon ID 1868215), whilst filtering out bovine sequences based on sequence similarity. We have let NCBI know about this misclassification.
Community Highlights
Pathoplexus is dedicated to serving the pathogen community, and as such we aim to be active participants in initiatives that help us connect with and better understand the needs of our users (and potential users!). Thus, we’re exceptionally pleased to announce that we are now an institutional member of the International Pathogen Surveillance Network (IPSN), convened by the WHO. We are excited to have the opportunity to connect with pathogen and public health researchers and workers worldwide and strengthen our ties with global partners in pathogen data infrastructure.
In addition, we’re grateful to the active pathogen communities for their work in improving standards and datasets. Recently, the mpox and RSV communities have been particularly active:
-
Newly designated mpox lineages are now annotated on Pathoplexus following community review
-
New RSV-B lineages have also been integrated, improving consistency and downstream analyses across datasets.
Citations
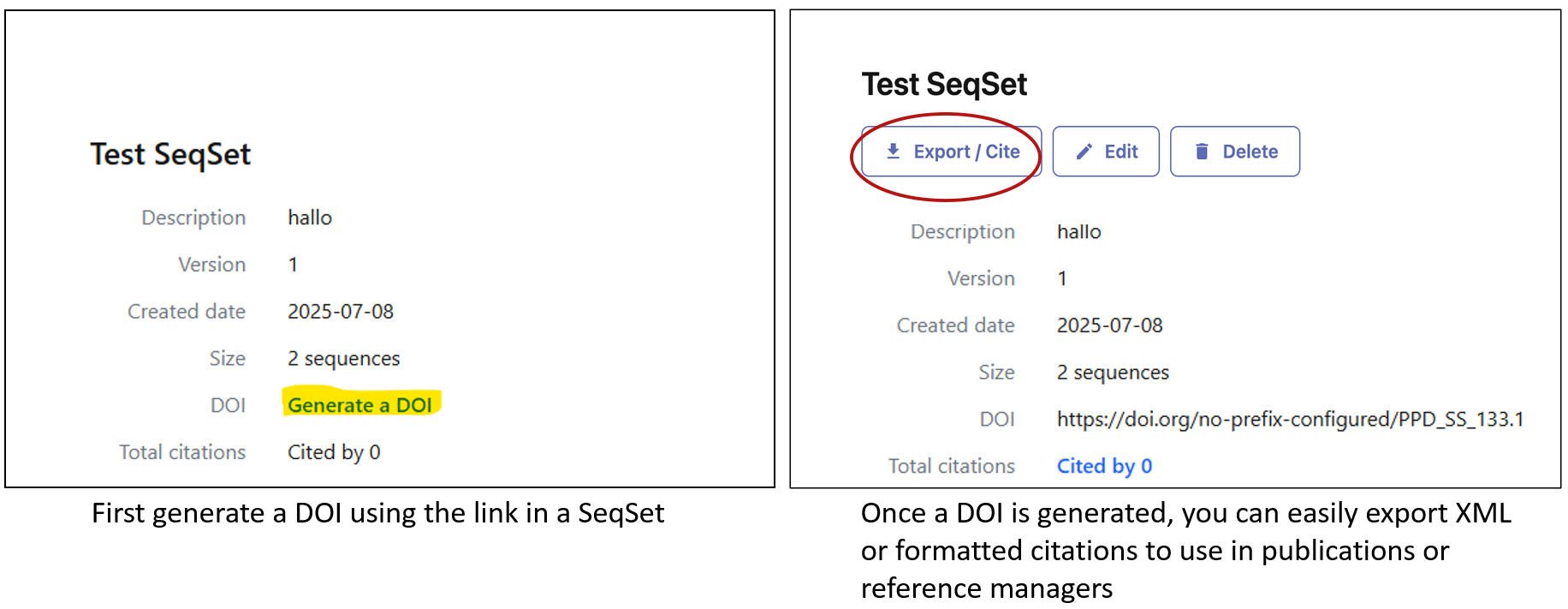
Additionally, we’d like to remind all those who use data from Pathoplexus that we do require use of Pathoplexus SeqSets and DOIs for citations in publications and preprints (this is especially critical when making use of Restricted-Use Data in any form). These DOIs should be cited as references to enable tracking; a citation-style reference can be generated using the ‘Export’ button at the top of the page.
Not only does proper citation ensure submitters receive full credit, but it also helps trace data provenance and use through time. Soon, this will allow submitters to see the total contribution their sequences have made towards work outside their own publications!
Technical Improvements
A series of backend and frontend enhancements have rolled out this quarter to make Pathoplexus faster, clearer, and more user-friendly:
Submission:
-
Accented characters are now supported in author names. Note that the INSDC accepts only ASCII characters. When submitting sequences to ENA, we hence replace non-ASCII characters with their nearest ASCII equivalent.
-
For users uploading measles sequences, we have a new field
means2idthat can be used to link sequences to the MeaNS2 database, if the sequences are also uploaded there, aiding with de-duplication. -
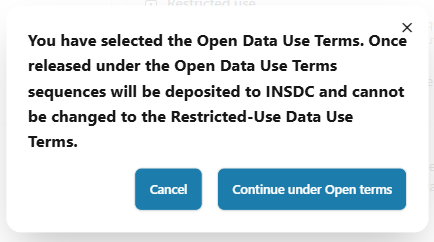
When submitting data to Pathoplexus as ‘Open,’ we now have a very clear pop-up to prevent submission without accidentally forgetting to change the Data Use Terms
Display:
-
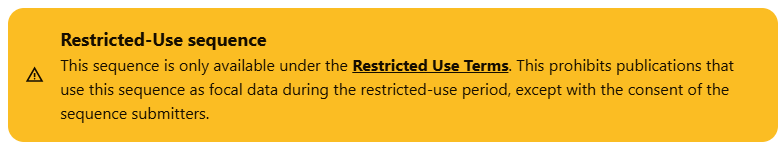
Improved banners on Restricted-Use Data sequence pages now provide more detail about usage terms
-
Group pages now show uploaded sequences (for logged-in users), giving users a clearer view of the data that’s been shared by a particular group - check it out on the Institut National de Recherche Biomédicale Group Page (you’ll need to log-in first!)

Access & Analysis:
-
FASTA downloads with rich headers are now 20× faster and can be compressed, reducing transfer time for large datasets.
-
Many search fields now support multiple-field search - allowing you to look at multiple countries, groups, and more at once!

-
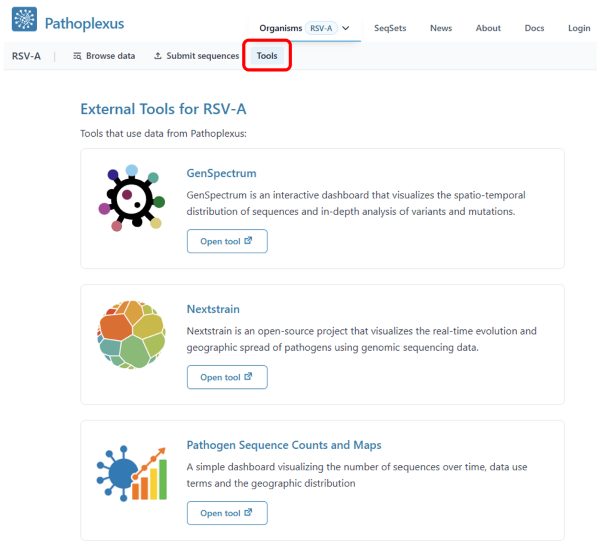
You can now view a list of external tools that make use of Pathoplexus data on each organism’s ‘Tools’ page. Interested in listing your own tool? We welcome integrating additional tools! Please open an issue to let us know! (Tools can also be for just one or a selection of pathogens)
Talks, Tutorials, and Media
Past talks
Pathoplexus and Loculus have been on the road - or rather, on the conference circuit!
-
The Binfie Podcast released a two-part feature on Pathoplexus and Loculus - covering their design philosophy, technical underpinnings, and vision for open pathogen data. Have a listen for yourself: Part I, Part II
-
The Loculus tutorial and talk at BC2 by Chaoran Chen and Anna Parker drew strong engagement, with many participants expressing interest in federated deployments.
-
Theo Sanderson presented Pathoplexus at IMMEM, highlighting how open-source infrastructure and transparent governance can power equitable data sharing.
-
Chaoran Chen presented Pathoplexus at the CERI Summer Scientific Seminar at Stellenbosch University.
-
Pathoplexus featured heavily at the PHA4GE-Conference pre-conference workshop and at the conferences itself! The well-attended workshop featured a diverse group getting to grips with how to create accounts, search Pathoplexus, and upload data - and was led by Arthur Shem Kasambula, Stephen Kanyerezi, Patricia Nabisubi (Uganda) and Emma Hodcroft and Chaoran Chen (Switzerland). The talk focused on exploring how open data ecosystems can serve public health. Here are a few photos of the coordinators and workshop in action!

-
Cornelius Roemer presented Pathoplexus at the 2025 ENA facilities on November 7.
Upcoming talks
- Emma Hodcroft will talk about Pathoplexus at the European Virus Bioinformatics Center “Viruses in Silico” lecture series on 27 Nov at 16.00 CET
Awards and Collaborations
We are incredibly pleased to announce that Loculus, the software powering Pathoplexus, was chosen as an SIB Remarkable Output 2024! In particular, judges commented, “Loculus is remarkable for enabling efficient and secure sharing of pathogen genomic data, which is crucial for public health responses to infectious diseases.”
In addition, the Loculus team is excited to be joining the Pathogen Data Network to aid more efficient and effective sharing of pathogen data. This initiative will help us expand and improve Loculus. Since Loculus is the software that underpins Pathoplexus - this means more features and benefits for Pathoplexus, as well!
New members
We’re pleased to welcome 8 new members to Pathoplexus this month: Vitor Borges (Portugal), Nick Goldman (UK), Arthur Shem Kasambula (Uganda), Stephen Kanyerezi (Uganda), Alejandra Gonzalez Sanchez (Spain), Laura Fahey (Ireland), Alan Rice (Ireland), and Senjuti Saha (Bangladesh).
Meetings, Minutes, and Resolutions
- Remember that you can always check our minutes from General Assemblies and Executive Board meetings, as well as Executive Board resolutions