News > Pathoplexus Turns One – and Adds Measles Virus!
Pathoplexus Turns One – and Adds Measles Virus!
By the Pathoplexus Team - 27 August 2025
Today we celebrate the first anniversary of Pathoplexus! 🎉🎂
Since launching on 27 August 2024 with four pathogens – Ebolavirus Sudan, Ebolavirus Zaire, West Nile virus, and Crimean-Congo haemorrhagic fever virus – we’ve grown further into the truly global, community-driven platform for transparent, equitable, and impactful pathogen sequence sharing that we strive to be.
A Year of milestones
Over the past 12 months, together with our community, we have:
-
Expanded our supported pathogens - adding mpox in December 2024, and RSV & HMPV in May 2025, in response to community requests.
-
Enabled rapid, global sharing - receiving direct submissions from 5 continents, including more mpox sequences in 2025 than any other database, and large RSV datasets from international collaborations.
-
Introduced major software improvements - from Excel metadata uploads and advanced search qualifiers, to integrated “link-out” tools for quick phylogenetic and geographic context.
-
Showed the importance of protected sequence sharing - enabling rapid public health use while protecting publication rights, with uptake from partners in Africa and South America.
-
Engaged widely - presenting Pathoplexus at WHO IPSN events, major scientific conferences, and workshops on data sharing, as well as featuring in Lancet Microbe, Science, Nature, and national media.
-
Received recognition - including the 2024 Swiss National Open Research Data Prize for our commitment to openness and community governance.
All of this has been achieved while being powered largely by volunteer effort - a testament to the dedication of our development team, members, curators, and Executive Board.
Welcoming Measles Virus to Pathoplexus
We’re excited to mark our first birthday by adding measles virus (MeV) as our newest supported pathogen!
Following strong community support and an Executive Board resolution, Pathoplexus now offers streamlined, user-friendly MeV data submission, with the same sharing options as our other pathogens:
- Open - sequences are shared immediately with INSDC-member databases.
- Restricted-Use - sequences can be shared rapidly for public health use while reserving publication rights for up to 1 year.
Adding measles virus comes at a critical time:
- Measles cases have surged dramatically in the US this year, making real-time genomic tracking vital in North America.
- Global travel and declining vaccination rates in some regions are increasing the risk of further outbreaks worldwide.
- More labs than ever are generating full-genome MeV sequences, offering opportunities for deeper insights into transmission, introductions, and viral evolution.
We believe supporting rapid MeV sequence sharing will strengthen surveillance and research - and we’re eager to work with the measles community to make this happen.
Other Updates:
New uploads:
-
We were pleased to receive 256 new direct mpox sequence uploads, from National Public Health Agency (NPHA), Ministry of Health, and Kenema Government Hospital, Sierra Leone; National Health Agency (NPHA), Central Public Health Reference Laboratory, Institute de Pasteur de Dakar, Sierra Leone; Instituto Nacional de Saude Doutor Ricardo Jorge (INSA), Portugal; Helmholtz Institute for One Health, Greifswalt Germany and Centre Suisse de Recherches Scientifiques, Cote d’Ivoire; Cliniques Universitaires de Kinshasa and Institut National de Recherche Biomedicale, DRC; National Virus Reference Laboratory, University College Dublin, Ireland; and Institut Pasteur de Bangui, Central African Republic
-
For West Nile, we received 308 sequences from San Diego County Public Health Laboratory, California, USA and University of Nebraska Medical Center, USA
Grant success:
- We’re incredibly happy to announce a successful small grant from “ETH4D Research Challenge” as a partnership between ETH Zurich, Pathoplexus, and Stephen Kanyerezi in Uganda. The aim of ETH4D is to aid collaborations between ETH research groups, non-academic partners, and partners from low- and lower-middle income countries. Our grant will focus on further developing Pathoplexus and Loculus, creating workshops and training materials, gathering user feedback, and developing our community.
Technical improvements:
-
It’s now possible to add GISAID accessions to sequences uploaded to Pathoplexus to aid de-duplication efforts. Simply add the accession (either on initial upload, or in a revision later) to the
gisaidIsolateIdmetadata field. (This is only available on organisms supported by GISAID.) -
Bulk selection made easier: You can now quickly select multiple sequences on the search page by clicking and dragging your mouse over the checkboxes —- no need to click each one individually.
-
Links to sequences at any INSDC database: Pathoplexus now offers a dropdown menu on our links to INSDC accessions, allowing you to view sequences in your preferred INSDC partner database—whether that’s GenBank, ENA, or DDBJ—making it easier to work with the platform you’re most familiar with.
-
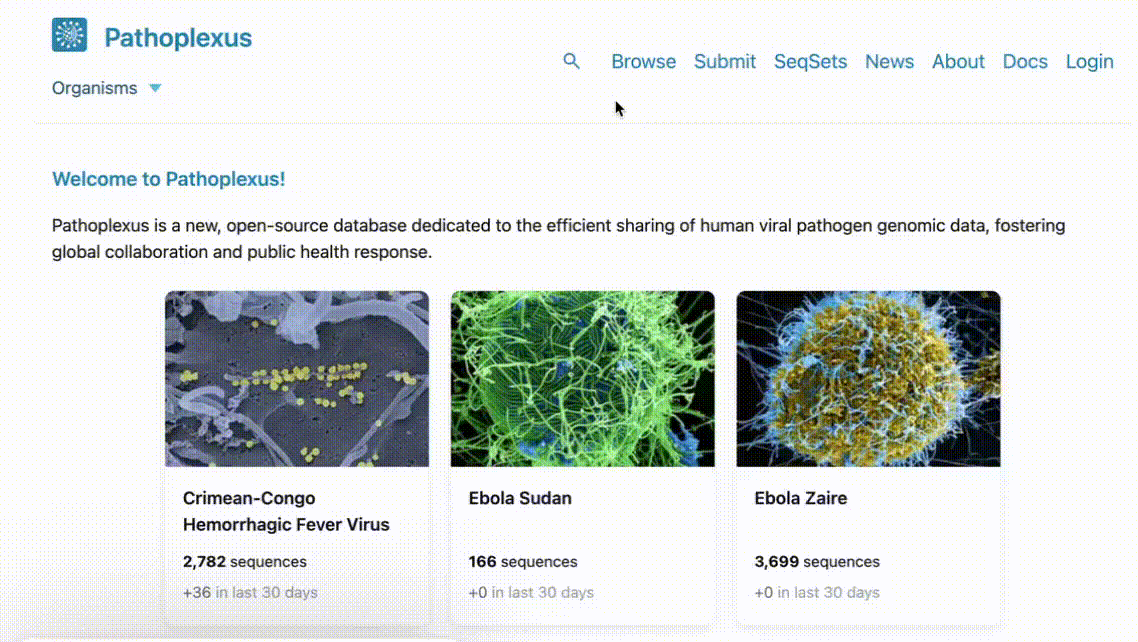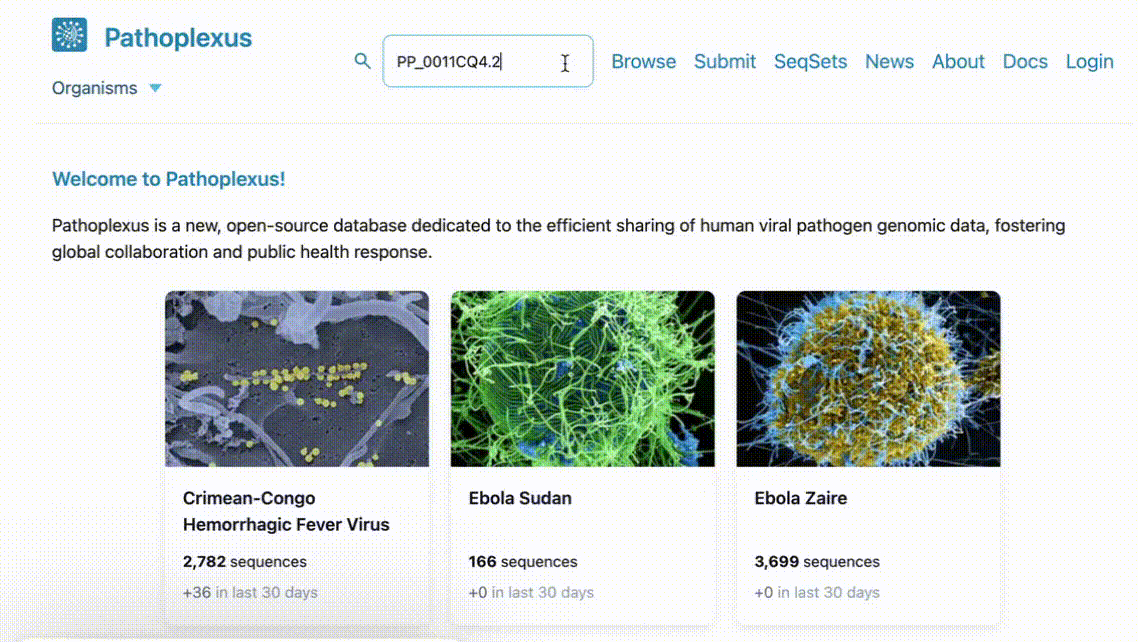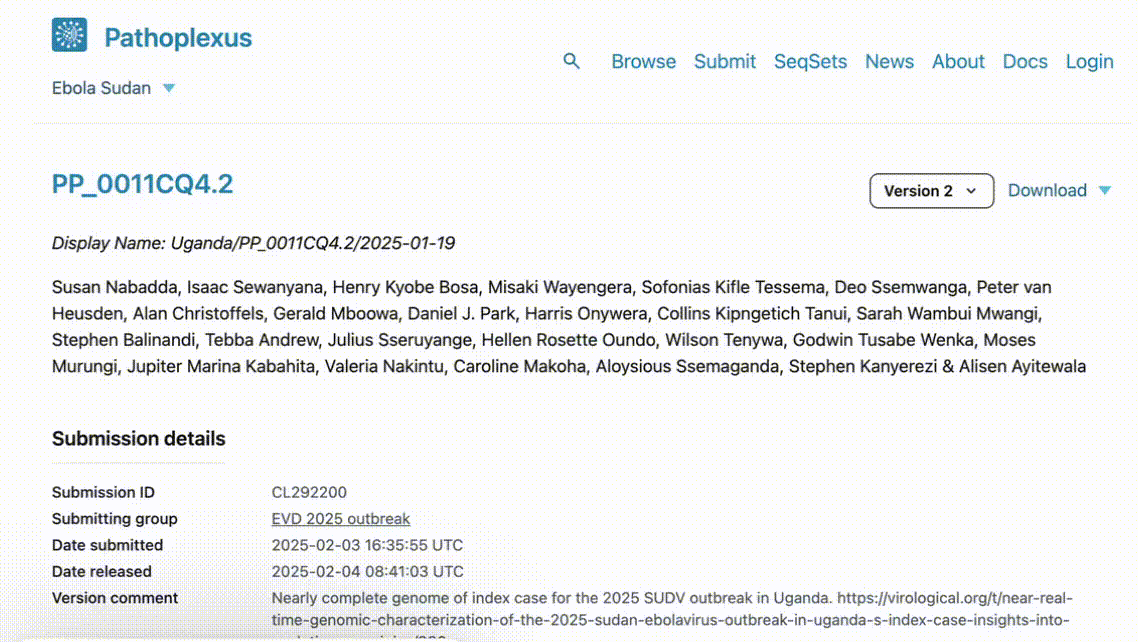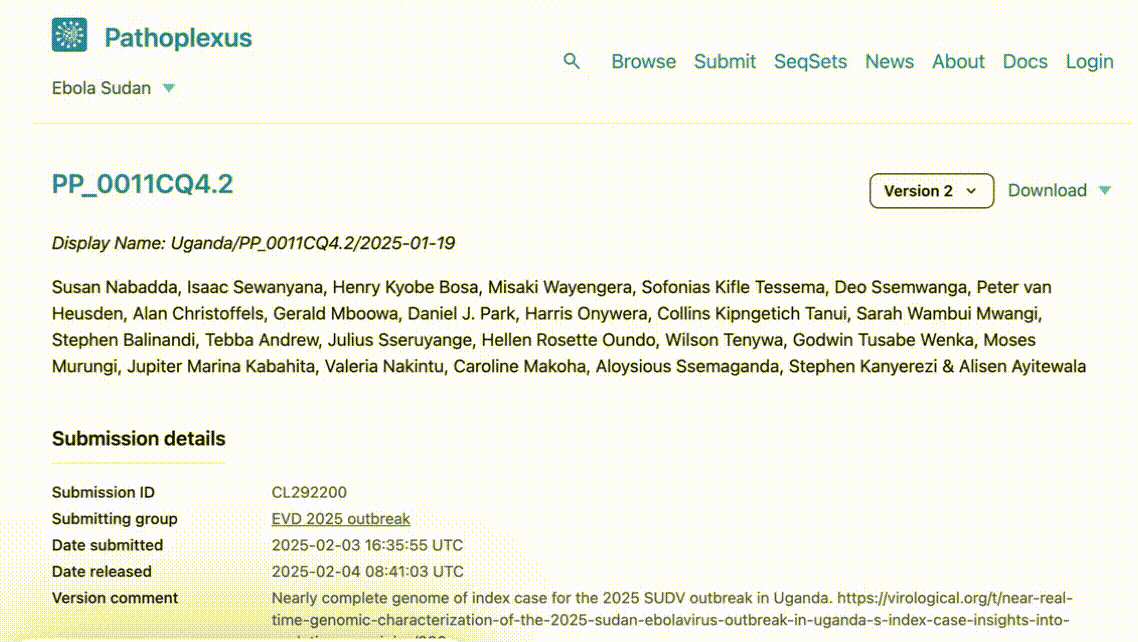
Find a sequence faster than ever by using our new expanding search icon at the top of the front page - just paste in a Pathoplexus accession and go straight there!
You can always keep track of all the latest changes to the software that powers Pathoplexus in the Loculus repository.
We are incredibly grateful to everyone who has contributed data, feedback, development, curation, and advocacy over the past year. Pathoplexus exists for - and because of - the global community working to understand and respond to viruses of public health importance.
Here’s to our second year, and to building an even more connected, equitable future for pathogen genomics! 🎊
🔗 Explore measles virus data on Pathoplexus: pathoplexus.org/measles/search
🔗 See all supported pathogens: pathoplexus.org
🔗 Learn how to submit your data: pathoplexus.org/docs/how-to/upload-sequences